
Rev. Fac. Agron. (LUZ). 1998, 15: 69-86
Microbial interactions in the rumen
Interacciones entre los microrganismos ruminales
Recibido el 07-09-1997l Aceptado el
10-12-1997
Department of Animal Sciences Ohio Agricultural Research and Development Center The Ohio
State University, Wooster, OH 44691-4096.
Burk A. Dehority
Abstract
Key words: rumen microorganism, interactions.
Resumen
Palabras claves: microorganismos ruminales, interacciones.
Introduction
The rumen is a large pouch in the foregut of numerous herbivores, which acts as a storage area in which ingested food is fermented by a complex, anaerobic microbial population. This population consists of about 1010 bacteria, 106 ciliate protozoa, and 106 phycomycete fungi per ml, which ferment the diet to volatile fatty acids, microbial protein and vitamins. The establishment and maintenance of a stable population is dependent upon diet, level of feeding, frequency of feeding and microbial interactions. Effects of diet, level of feeding and frequency of feeding have been studied rather extensively (19); however, information about microbial interactions is quite limited. Much of our knowledge about interactions is based primarily on observations or in vitro studies. Thus, understanding the relationship of these studies to the rumen itself is also a little uncertain. One might expect that microbial interactions can be relatively subtle and their effects to be quite com plex.
In general, interactions can be either positive or negative and can occur both within and between microbial types. Prins and Vorstenbosch (51) have suggested that the associations between the different microorganisms can be described by three terms: mutualism, an association which is beneficial to both; commensalism, an association which is beneficial to one of the partners but without effect on the other; and parasitism, an association in which one of the partners gains at the expense of the other. Figura 1 presents a diagram which outlines the various interactions which have been observed to date. The only positive interactions would be synergism between fungi and bacteria and synergism and cross feeding between bacterial species. These would fit under the categories of both mutualism and commensalism. All other effects listed are negative or parasitic in nature.
Protozoal interactions
Between protozoa. Probably the best known interaction between rumen protozoa is predation of one ciliate species by another. Lubinsky (40) reported observing numerous cases of protozoal predation in his studies of Canadian reindeer and domestic ruminants in Punjab, India. He considered the predation to be accidental and occur primarily in species with a larger body-size. Williams and Coleman (61) have summarized the reported obser vations of apparently accidental predation by various authors, identifying both the predator and prey species. In contrast, several examples of specific predation have been observed, i.e., differing from accidental predation in that it leads to complete removal from the population of those species which form the prey. Eadie (22) first noted that two general types of rumen ciliate populations seemed to occur. Essentially, Type A contained entodinia, isotrichids and Polyplastron multivesiculatum, whereas type B contained entodinia and isotrichids along with a larger entodiniomorph, Eudiplodinium and/or Epidinium. Type A was most prevalent in sheep and Type B was most frequent in cattle. However, cross-inoculation of the two types always resulted in an irreversible change to the type A population. In subsequent studies (23) was able to demonstrate that predation by Polyplastron appeared to be the major means by which this organism and the type A fauna predominates. Starvation of Polyplas-tron seemed to prevent or decrease predation rather than stimulate this activity; however, it did result in a slight increase in cannibalism. Polyplastron will eliminate Epidinium, along with many Eudiplodinium and Ostracodinium species. Several years ago, new lambs were brought into the author's barn for research studies. Observation of their fauna indicated the presence of a type B population, containing Epidinium and no Polyplastron. When these animals were inoculated with lamb rumen contents containing Polyplastron, a rapid decline and disappearance of Epidinium was observed, with a concomitant establishment of Polyplastron. Coleman and coworkers (13, 14) found that Polyplastron requires the presence of Epidinium for growth in vitro, which they engulf and appear to use as a food source. Several other species of Diplodiniinae could replace Epidinium, but Entodinia could not. This same group also found that for growth in vitro, Entodinium bursa has an absolute requirement for the spineless form of Entodinium caudatum. E. bursa apparently engulfs E. caudatum posterior end first, and the spines inhibit or slow down their engulfment (62). All other species of Entodinium were unable to support growth of E. bursa.
The other major interaction between protozoa was also described by Eadie (23). She observed that Epidinium consistently predominates over Ophryoscolex when the two are mixed in vivo. However, since no predation could be observed, she suggested that other factors such as nutrient or food competition could be responsible.
Between bacteria and protozoa. Predation of rumen bacteria by the rumen ciliate protozoa was first recognized by Gutierrez and coworkers (29, 30, 31). In an extensive series of studies, Coleman and his group have demonstrated the rapid engulfment of bacteria by the rumen protozoa (11). Although all the ophryoscolecid protozoa tested have some ability to take up amino acids from the medium, they cannot be cultured in vitro in the absence of bacteria which appear to be their major source of nitrogenous compounds (12). However, utilization of the bacterial digestion products is rather inefficient, with considerable quantities, up to 50%, of the amino acids being released back into the rumen (11). It should be noted that the bacteria apparently also provide other required nutrients for protozoal growth (47). As a result of protozoal predation, bacterial concentrations are lower in rumen contents of animals with ciliate protozoa, and concentrations increase when the animals are defaunated (61). It has been suggested that both the rate and efficiency of bacterial growth can be expected to increase as a result of protozoal predation, primarily because more food and nutrients are available (50). In most cases, predation on bacteria by the rumen ciliates does not appear to be species-specific, but rather random and more a function of bacterial concentration (50). One possible exception might be that the bacteria attached to fiber (cellulolytic and hemicellulolytic species) may be less likely to be ingested.
Based on the fact that to date we have been unable to establish axenic bacterial-free cultures of the rumen protozoa, the protozoa must be considered as parasitic. Although the bacteria can function in the absence of the protozoa, the converse does not appear to be true.
Between protozoa and fungi. Most of the evidence for predation of fungi by the protozoa is circumstantial, based on an increase in fungal concentrations when animals are defaunated (48, 53, 58). However, in other studies fungal concentrations were not increased in defaunated sheep (44, 60). Williams and Withers (63) did not observe a decrease in fungal concentrations when defaunated sheep were refaunated. In most of the above listed studies, the animals were fed high roughage diets and fungal concentrations were determined in rumen fluid using either the roll tube procedure of Joblin or direct microscopic counts (48). Bond (7) used the MPN procedure of Obispo and Dehority to measure fungal concentrations in whole rumen contents of three sheep before and after defaunation. No effect was observed in two of the sheep, while a 10-fold increase in fungal concentrations occurred in the third animal.
Other observations which might bear on this subject would be: (1) scanning electron micrographs which show protozoa ingesting fungal rhizoids and sporangia (62, 64); and (2) turnover of fungal protein is decreased in in vitro fermentations with rumen fluid from defaunated sheep (44). It should be noted, that in general, increases in concentration and decreases in protein turnover in defaunated animals were much greater for bacteria than fungi. Williams et al. (1994) subsequently demonstrated an increase in the breakdown of fungi in vitro when incubated with various species of protozoa. Although the overall evidence for predation of fungi by protozoa is somewhat variable, it does suggest that predation occurs; however, probably to a lesser extent than with the bacteria.
Bacterial interactions
Between bacteria. As shown in figure 1, interactions between bacterial species can be both positive, i.e., synergism and crossfeeding or negative, i.e., production of compounds in hibitory to other bacterial species.
Positive effects. The most obvious interactions which have been observed between different species of rumen bacteria is the marked synergism in digestion of structural carbohydrates. Table 1 presents the mean extent of cellulose digestion from 12 forages by pure cultures of rumen bacteria (20). Fermentations were run with each strain alone and in all possible combinations of two. The major cellu lolytic species, Fibrobacter succinogenes (A3c), Ruminococcus albus (7) and Ruminococcus flavefaciens, (B34b and B1a) all digested considerable amounts of cellulose alone and no increases were observed when any two were combined in the same fermentation. The noncellulolytic organism, Prevotella ruminicola (H8a) did not digest any cellulose from the forages; however, when combined with any of the cellulolytic species, cellulose digestion was increased. None of the combinations with the weakly cellulolytic species, Butyrivibrio fibrisolvens H10b, increased cellulose digestion.
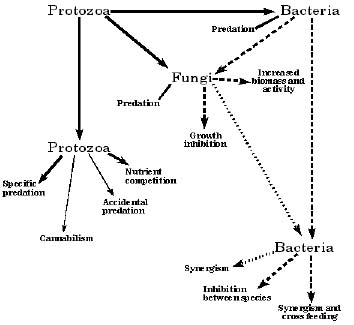
Figure 1. Diagrammatic sketch of microbial interactions which occur in the rumen. Protozoal effects are designated by solid arrow lines; bacterial effects by the long dashes and fungal effects by the short dashes. Effects in smaller type are minor in scope. Those effects inside of boxes are positive in nature, while all others are negative.
Table 1. Extent of forage cellulose digestion by pure cultures of rumen cellulolytic bacteria singly and in all combinations of two.*
| Cellulose digestion, %‡ | ||||||
| Organism 2 | ||||||
| Organism 1† | A3c | 7 | B34b | B1a | H10b | H8a |
| A3c | 61.9 | 63.1 | 44.7a | 62.2 | 63.5 | 66.2a |
| 7 | 44.4 | 41.2 | 39.9b | 40.3b | 48.8a | |
| B34b | 44.1 | 43.5 | 46.1 | 47.0b | ||
| B1a | 36.3 | 32.1b | 42.2a | |||
| H10b | 8.7 | 6.1 | ||||
| H8a | 1.6 | |||||
*Data from Dehority and Scott (20). Values are the mean of twelve forages (eight grass and four alfalfa samples). †A3c, Fibrobacter succinogenes; 7, Ruminococcus albus; B34b and B1a, Ruminococcus flavefaciens; H8a, Prevotella ruminicola. ‡Within a given row, a indicates a difference at P < .01 and b at P < .05, with respect to the mean cellulose digestibility for that bacterial strain alone.
Table. 2. Percent degradation (Deg.) and utilization (Utl.) of hemicellulose from alfalfa (Medicago sativa), fescue grass (Festuca pratensis) and isolated fescue grass hemicellulose*
| Substrate | ||||||
| Alfalfa | Fescue | Isolated fescue hemicellulose‡ |
||||
| Organism† | Deg. | Utl. | Deg. | Utl. | Deg. | Utl |
| B34b | 56.3 | 2.1 | 66.6 | 3.0 | 88.5 | 0 |
| H10b | 35.4 | 34.1 | 44.8 | 38.0 | 87.5 | 83.8 |
| H8a | 33.6 | 33.9 | 2.7 | 2.0 | 82.0 | 80.4 |
| D15d | 49.5 | 23.2 | 4.0 | 1.3 | 1.7 | 1.7 |
| B34b+H10b | 61.9 | 43.2 | 67.3 | 64.8 | 91.3 | 87.8 |
| B34b+H8a | 59.6 | 54.8 | 69.0 | 67.7 | 93.9 | 87.0 |
| B34b+D15d | 61.8 | 14.9 | 67.9 | 3.9 | 87.0 | 3.8 |
| All | 61.8 | 58.4 | 67.6 | 65.9 | 87.4 | 85.7 |
*Values from Coen and Dehority (10). †B34b, Ruminococcus flavefaciens; H10b, Butyrivibrio fibrisolvens; H8a, Prevotella ruminicola; D15d, Lachnospira multiparus. ‡Hemicellulose was isolated from the same stand of fescue grass.
Even more marked synergism between bacterial species has been noted in the digestion of forage hemicelluloses. Dehority (15) observed that many of the cellulolytic species were able to degrade (change to a form soluble in acidified 80% ethanol) isolated hemicelluloses regardless of their ability to utilize them as energy sources. The hemicellulolytic species extensively utilized these substrates as a source of energy. In a subsequent study, Coen and Dehority (10) found that many of the hemicellulose utilizing species were unable to degrade and utilize the hemicelluloses from intact forages, especially from grasses. However, if the hemicellulose was physically isolated from the grass it was almost completely degraded by the cellulolytic species or degraded and utilized by the hemicellulolytic species (table 2). Thus, combining a hemicellulose degrading but nonutilizing cellulolytic species with a nondegrading but utilizing hemicellulolytic species resulted in extensive hemicellulose digestion from the intact forage. These findings were confirmed and expanded in later studies by Morris and van Gylswyk (42), Chesson et al. (9) and Osborne and Dehority (49). Dehority (18) has compiled a list of those rumen bacteria which can degrade hemicel lulose (depolymerase activity), utilize hemicellulose (glycosidose activity) or both.
In a recent study, Fondevila and Dehority (25) developed procedures for sequential addition of organisms as a means to study the synergism in forage hemicellulose digestion. One organism was allowed to ferment a forage substrate, after which the culture tube was sterilized and then inoculated with a second organism. Using Fibrobacter succinogenes A3c, Prevotella ruminicola H2b and Ruminococcus flavefaciens B34b, singly and in all possible combinations, hemicellulose utilization was increased by all combinations of the cellulolytic species (A3c or B34b) with the nonce-llulolytic, hemicellulolytic H2b. The effect of sequential addition of the two organisms is shown in table 3. In general, adding either of the cellulolytic species first gave utilization values similar to those when both species were added at the same time. However, adding the hemicellulolytic organism first markedly reduced the extent of utilization. These data clearly fit the model of the cellulolytic species degrading or solubilizing the hemicellulose so that it can be utilized by the limited degrading hemicellulose utilizer.
Gradel and Dehority (28) subsequently found that several species of cellulolytic bacteria possessed pectin dipolymerase activity, but not the enzymatic capabilities to utilize the resulting oligogalacturoides or galactuoric acid as an energy source. This activity was also confirmed later by Morris and van Gylswyk (42). These characteristics were quite similar to those previously observed with regard to hemicellulose digestion, and as might be expected, combining a cellulolytic and purified pectin utilizing species resulted in an increase in forage pectin utilization (28). In a later study on forage pectin digestion, Osborne and Dehority (49) obtained some surprising results in that a marked synergism resulted from the combination of F. succinogenes A3c and P. ruminicola H2b (table 4). Neither of these organisms had previously shown much activity against purified pectin (15); however, A3c degraded and H2b utilized forage pectin quite extensively. Conversely, D15d extensively degraded and utilized purified pectin, but had little activity against intact forage pectin. Thus, isolation and char acterization of rumen bacteria on purified polysaccharides can be misleading with respect to their activities in fermenting these substrates from intact forages.
Table 3. Percent degradation (Deg.) and utilization (Util.) of hemicellulose from intact forage by two pure cultures of rumen bacteria added together or sequentially*
| Organism† | Forage | ||||
| Orchardgrass | Alfalfa | ||||
| First | Second | Deg. | Utl. | Deg. | Utl. |
| A3c+H2b | 60.7a | 58.7a | 40.1 | 30.7a | |
| A3c | H2b | 61.5a | 57.0a | 43.8 | 28.0a |
| H2b | A3c | 44.9b | 20.4b | 37.6 | 14.1b |
| B4b+H2b | 40.2 | 36.0a | 41.4 | 31.5a | |
| B34b | H2b | 37.0 | 34.5a | 35.9 | 24.8a |
| H2b | B34b | 39.9 | 26.0b | 44.6 | 14.6b |
| A3c+B34b | 45.9a | 21.1 | 43.6 | 18.9 | |
| A3c | B34b | 60.6b | 27.6 | 51.8 | 21.4 |
| B34b | A3c | 35.6c | 22.0 | 47.8 | 23.4 |
*Data from Fondevila and Dehority (25). † A3c, Fibrobacter succinogenes; H2b, Prevotella ruminicola; B34b, Ruminococcus flavefaciens. a,b,c For each pair of organisms, means in the same column followed by different superscripts are different at P < .05.
Crossfeeding of hydrolysis products, utilization of end-products or production of an essential nutrient are the other types of positive interactions which can occur between bacterial species. For example, non-cellulolytic bacteria can utilize the cello dextrins produced by the cellulolytic species (54). Rumen methanogens obtain energy by converting the metabolic end-products hydrogen and carbon dioxide to methane (55, 66). Williams et al. (64) observed an increase in xylan utilization when R. flavefaciens was cocultured with Methanobrevibacter smithii, with the fermentation becoming acetogenic. Conversion of succinate, a normal end product of several cellulolytic and amylolytic bacteria, to propionate, is another example of this type of synergism between species (57, 66). Production of a nutrient by one bacterial species which is essential for the growth of a second species, also occurs in the rumen. Generally the nutrients involved are either vitamins, amino acids or branched-chain fatty acids (41, 66).
Table 4. Percent degradation (Deg.) and utilization (Utl.) of pectin by pure cultures of rumen bacteria, singly and in all combinations*
| Immature orchardgrass | Purified pectin | |||
| Organism† | Deg. | Utl. | Deg. | Utl. |
| A3c | 68.5ac | 0.0a | 17.9a | 9.5a |
| H2b | 54.9a | 46.1b | 12.1b | 5.1a |
| D15d | 18.9b | 6.8a | 87.1c | 73.2b |
| A3c + H2b | 83.9d | 75.3c | 17.9a | 8.1a |
| A3c + D15d | 78.3cd | 0.0a | 87.8c | 73.2b |
| H2b + D15d | 56.6a | 49.4b | 87.9c | 73.4b |
*Data from Osborne and Dehority (49). †A3c, Fibrobacter succinogenes; H2b, Prevotella ruminicola, D15d, Lachnospira multiparus. a,b,c,d Means in the same column followed by different superscripts differ at P < .05).
Synergism which results when one bacterial species "unmasks" or makes a substrate available to a second organism, crossfeeding, use of end products and nutrient production, can all be classified under commensalism. That is, the second species benefits from the action of the first, without any detrimental effect on the first organism.
Negative effects. In several of the studies cited earlier on synergisms between bacterial species in digestion of forage structural carbohydrates, it was also observed that some combinations reduced the extent of digestion. In table 1, forage cellulose digestion was decreased by combining F. succinogenes A3c with R. flavefaciens B34b, or R. albus 7 with R. flavefaciens B1a. These decreases in intact forage degradation have subsequently been observed with other strains of these species, i.e., between F. succinogenes and R. flavefaciens (56) and between R. albus and R. flavefaciens (45). Similar decreases have also been observed in both hemicellulose utilization (10) and pectin utilization (28). One possible explanation for these negative responses would be that the two organisms produce different depolymerases, which act at different sites on the polysaccharide. Different oligosaccharides are then produced which cannot be further metabolized by the available glycosidases. An additional possibility was suggested by the studies of Odenyo et al. (45). They reported that R. albus 8 produced proteinaceous factors which inhibited the growth of R. flavefaciens FD-1; but not F. succinogenes S85. They suggested that this inhibitory compound was a bacteriocin-like substance. Bacteriocins are bactericidal proteins produced by species which are generally inhibitory to other species closely related to the producer. Production and properties of bacterocins have been studied quite extensively among bacteria used in fermenting dairy products (3).
While trying to develop a selective medium for enumeration of R. albus 7 and R. flavefaciens B1a in coculture, Chan and Dehority (8) observed that growth of R. flavefaciens was inhibited when the cultures were mixed. R. albus 7 was found to produce an inhibitory substance that was present in cell-free culture filtrates, was heat-labile and destroyed by a proteolytic enzyme. R. albus 7 plus two additional strains of R. albus all produced inhibitory activity against B1a and several additional R. flavefaciens strains, but not against F. succinogenes, B. fibrisolvens or P. ruminicola. These data support the previous observations by Odenyo et al. (45), that R. albus produced a bacteriocin-like substance which is inhibitory to many strains of the closely related species R. flavefaciens.
Kalmokoff and Teather (37) screened 49 Butyrivibrio fibrisolvens isolates for bacteriocin production. They found that twenty five produced products which showed varying degrees of inhibition to the other isolates plus some unrelated Gram-positive rumen bacteria. The inhibitory activity from 18 of the 25 strains was sensitive to proteolytic activity.
An apparently different type of antagonism was observed by Fondevila and Dehority (26) who used sequential addition experiments to study the antagonism between R. flavefaciens B34b and F. succinogenes A3c (table 1). They found that combining these two organisms markedly reduced forage cellulose digestion from that of A3c alone. When the two cultures were added sequentially, cellulose digestion was not different from A3c alone, regardless of the order in which the cultures were added. Table 5 presents data from an additional experiment by these authors, which clearly indicates that R. flavefaciens only suppresses the cellulolytic activity or growth of F. succinogenes when the organisms are simultaneously present in the fermentation medium. It was also of interest that autoclaving at 121°C for 20 min did not destroy the inhibitory material.
Between bacteria and fungi
Positive effects. Since the rumen fungi produce appreciably quantities of hydrogen, they can interact with hydrogen utilizing organisms which in turn alters their metabolite production. Methanogens are the principal hydrogen utilizers in the rumen, and stable cocultures of fungi and methanogens have been successfully established in vitro (4, 43; 27). In pure culture, the fungi produce acetic acid, lactic acid, formic acid, ethanol, carbon dioxide and hydrogen. In the presence of methanogens, the fermentation becomes acetogenic, i.e., acetic acid production increases, lactic acid and ethanol formation decreases and hydrogen and formic acid to not accumulate. This alteration in metabolism results in increased energy per mole of hexose fermented and a measurable increase in fungal biomass (64). The rate and extent of cellulose digestion from filter paper increases in cocultures of fungi and methanogens (4, 65). Similar increases were noted in hemicellulose utilization with the fungal-methanogen cocultures (36). These increases in utilization are generally attributed to removal of metabolites which inhibit fungal growth. However, increases in degradation of these polysaccharides from intact plant tissue by cocultures is considerably less, perhaps because of restricted accessibility of the substrate (35, 64). Extent of the increase in cell wall digestibility by cocultures varies markedly with the strain of fungus and species of methanogen.
Table 5. Percent digestion of cellulose from intact orchardgrass by F. succinogenes and R. flavefaciens, alone, in coculture or added sequentially*
| Organism† | ||
| First | Second | Cellulose digestion, % |
| A3c | None | 48.9a |
| B34b | None | 29.8b |
| A3c + B34b | None | 29.7b |
| B34b | A3c | 43.3a |
| B34b | A3c + B34b | 29.6b |
| A3c + B34b | A3c | 29.7b |
| A3c + B34b | A3c + B34b | 29.9b |
*Data from Fondevila and Dehority (26). † A3c, Fibrobacter succinogenes; B34b, Ruminococcus flavefaciens. a,bMeans in the column followed by different superscripts differ at P < .05.
The fungi also are involved in cross feeding in that they release free sugars, which in addition to several of their normal metabolites, except acetate, serve as energy sources for other bacterial species. The fungi themselves also may depend on the bacteria to supply their nutritional requirements of B vitamins, heme, amino acids, etc. (64).
Negative effects. Preliminary studies by Lowe et al. (39) and Akin and Windham (1) suggested that rumen fluid or rumen bacteria could inhibit fungal growth and activity. More recently, negative or inhibitory effects on fungal cellulose digestion were observed when the fungi are cocultured with Ruminococcus species (5, 6, 33, 52, 59). No such inhibitory activity was observed with F. succinogenes cocul-tures. Stewart et al. (59) found that the inhibitory compound was present in cell-free culture filtrates of Ruminococcus, could be destroyed by autoclaving and was protein in nature. Inhibition was not observed when the fungi were grown on glucose, which suggested an interference with attachment of the fungi to an insoluble substrate. These observations were later confirmed by Bernalier et al. (6).
Dehority and Tirabasso (21) measured cellulose digestion along with fungal and bacterial numbers in vitro using rumen fluid as an inoculum. Cellulose digestion and changes in microbial concentrations during the fermentation of purified cellulose by rumen contents, with and without added antibiotics are shown in table 6. It is quite obvious from these data that the fungi do not grow unless bacterial growth is suppressed with antibiotics. Similar results were obtained using intact alfalfa as substrate. The normal bacterial fermentation products do not appear to be responsible for this degree of inhibition (34). Subsequent studies by Dehority and Tirabasso (21) indicated that an inhibitory factor was produced in vitro by rumen bacteria which was also present in rumen fluid. This inhibitory activity is stable to autoclaving and not degraded by proteolytic enzymes (Dehority and Tirabasso, unpublished). Thus, the inhibitory factor or factors in these studies is apparently different from that previously observed in the pure culture studies with Ruminococcus.
Fungal interactions
Most of the interactions between the fungi, bacteria and protozoa have been discussed in the previous sections. However, one additional interaction concerns the potential ability of the fungi to physically weaken and disrupt the physical structure of intact forages. Ho et al. (32) have described the growth of appressorium-like structures at those sites where rumen fungal rhizoids come into contact with intact rigid cell walls. At the point of contact with the cell wall, the appressorium produced a fine penetration peg which penetrated the cell wall and continued to grow and elongate, producing normal rhizoids. These rhizoids in turn formed appresoria where they came into contact with walls of the adjacent cells. This process could be of importance in the digestion of intact forages, allowing the bacteria access to the structural polysaccharides. Engles and Brice (24) have observed the presence of a layer which lines the inner surface of lignified cell walls, restricting access of rumen microorganisms even after they have entered into the lumen of the cell. Akin et al. (2) measured a marked reduction in the textural strength of stem internodes in forages incubated with rumen fungi, as compared to incubation with mixed rumen bacteria. Thus, it appears that the fungi may act synergistically in conjunction with the bacteria, by physically disrupting the lignified forage cells. The rumen bacteria are thus able to enter into plant stems and tissues where the forage polysaccharide substrates are more accessible for digestion.
Table 6. Cellulose digestion and changes in microbial concentrations during the fermentation of purified cellulose by rumen contents with and without added antibiotics*
| Without antibiotics | With antibiotics | |||||
| Time | Cellulose | Bacteria† | Fungi† | Cellulose | Bacteria† | Fungi† |
| (h) | dig. % | (× 107) | (× 102) | dig. % | (× 107) | (× 102) |
| 0 | 0 | 9 | 12 | 0 | 9 | 12 |
| 24 | 38 | 1000 | 0.01 | 1 | 0.0004 | 16 |
| 30 | 51 | 451 | 0.02 | 3 | 0.002 | 50 |
| 48 | 57 | 290 | 0 | 17 | 0.004 | 230 |
| 72 | 70 | 38 | 0 | 47 | 0.003 | 510 |
*Data from Dehority and Tirabasso (21). †Concentration per ml of fermentation medium.
Conclusions
Most of the interactions observed between rumen organisms are based on in vitro experiments, using both pure and mixed cultures. Whether these same interactions occur in vivo is somewhat difficult to measure. Since the metabolic activities of all three types of rumen microorganisms are quite similar, it might be expected that another organism would take over any activity specifically reduced by inhibition of a particular organism. Other factors must also be considered, i.e., the type of forage or feed and its potential digestibility as well as its rate of passage through the rumen. Using some of the data presented in table 1 and from Dehority (17), it may be possible to gain some insight into how these interactions could effect in vivo digestibilities. Table 7 shows that the highest amount of cellulose is digested in vitro by F. succinogenes A3c and P. ruminicola H8a. Digestion is reduced by combining A3c with R. flavefaciens B34b. A combination of these three cultures plus three additional cultures does not digest as much cellulose as A3c alone. However, in vivo digestibility , as measured by sheep digestion trials, is less than the A3c + H8a combination, similar to A3c alone and greater than all the others. Thus, the marked reduction which occurs by combining A3c and B34b is partially alleviated with addition of the four additional cultures and almost completely disappears in vivo. Rate of passage through the rumen would be expected to result in a slightly lower extent of in vivo cellulose digestion.
In summary, although the microbial interactions outlined in this paper are demonstrable in vitro, their importance in vivo may be extremely limited. The overall rumen fermentation appears to be quite homeostatic, and is perhaps controlled to a greater extent by factors related to the feed an animal consumes rather than a specific microbial population.
Table 7. Comparison between the mean cellulose digestion for 12 forages determined in vitro by pure cultures (singly, in coculture and in a combination of six cultures) and in vivo by sheep digestibility trials*
| Inoculum | Cellulose digestion, % |
| Fibrobacter succinogenes | A3c 61.9a |
| Ruminococcus flavefaciens | B34b 44.1b |
| F. succinogenes A3c + R. flavefaciens | B34b 44.7b |
| F. succinogenes A3c + Prevotella ruminicola | H8a 66.2c |
| Combination of 6 cultures† | 54.6d |
| In vivo digestibility trials | 59.8a |
*Data from Dehority and Scott (20) and Dehority (17). † F. succinogenes A3c; R. flavefaciens B1a and B34b; R. albus 7; P. ruminicola H8a; and Butyrivibrio fibrisolvens H10b. a,b,c,dMeans in the column followed by different superscripts differ at P < .05.
Literature cited
1. Akin, D. E. and W. R. Windham. 1989. Influence of diet on rumen fungi, p 75-81. In J. V. Nolan, R. A. Leng and D. I. Demeyer (ed.), The Roles of Protozoa and Fungi in Ruminant Digestion. Penambul Books, Armidale NSW, Australia.
2. Akin, D. E., C. E. Lyon, W. R. Windham and L. L. Rigsby. 1989. Physical degradation of lignified stem tissues by ruminal fungi. Appl. Environ. Microbiol. 55:611-616.
3. Barefoot, S. F. and C. G. Nettles. 1992. Antibiosis revisited: bacteriocins produced by dairy starter cultures. J. Dairy Sci. 76:2366-2379.
4. Bauchop, T. and D. O. Mountfort. 1981. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl. Environ. Microbiol. 42:1103-1110.
5. Bernalier, A., G. Fonty, F. Bonnemoy and P. Gouet. 1992. Degradation and fermentation of cellulose by the rumen anaerobic fungi in axenic cultures or in association with cellulolytic bacteria. Curr. Microbiol. 25:143-148.
6. Bernalier, A., G. Fonty, F. Bonnemoy and P. Gouet. 1993. Inhibition of the cellulolytic activity of Neocallimastix frontalis by Ruminococcus flavefaciens. J. Gen. Microbiol. 139:873-880.
7. Bond, D. R. 1994. The effects of fiber source, defaunation, and antibiotic treatment on the anaerobic fungi population in vivo. M.S. Thesis. The Ohio State University, Columbus.
8. Chan, W. W. and B. A. Dehority. 1995. Growth inhibition of Ruminococcus flavefaciens by Ruminococcus albus. p. 14. In: 23rd Biennial Conference on Rumen Function. Chicago, IL. Nov. 14-16.
9. Chesson, A., C. S. Stewart, K. Dalgarno and T. P. King. 1986. Degradation of isolated grass mesophyll, epidermis and fibre cell walls in the rumen and by cellulolytic rumen bacteria in axenic culture. J. Appl. Bacteriol. 60:327-336.
10. Coen, J. A. and B. A. Dehority. 1970. Degradation and utilization of hemicellulose from intact forages by pure cultures of rumen bacteria. Appl. Microbiol. 20:362-368.
11. Coleman, G. S. 1975. The interrelationship between rumen ciliate protozoa and bacteria. p 149-164. In I. W. McDonald and A.C.I. Warner (ed.), Digestion and Metabolism in the Ruminant, University of New England Publishing Unit, Armidale, Australia.
12. Coleman, G. S. 1979. Rumen ciliate protozoa. p 381-408. In M. Levandowsky and S. H. Hutner (ed.), Biochemistry and Physiology of Protozoa, Academic Press, New York.
13. Coleman, G. S. and F. J. Hall. 1978. Digestion of Epidinium ecaudatum caudatum by the rumen ciliate Polyplastron multivesiculatum as shown by studies in the electron microscope. Society for General Microbiology Quarterly 6:29-30.
14. Coleman, G. S., J. I. Davies and M. A. Cash. 1972. The cultivation of the rumen ciliates Epidinium ecaudatum caudatum and Polyplastron multivesiculatum in vitro. J. Gen. Microbiol. 73:509-521.
15. Dehority, B. A. 1965. Degradation and utilization of isolated hemicellulose by pure culture of cellulolytic rumen bacteria. J. Bacteriol. 89:1515-1520.
16. Dehority, B. A. 1973. Hemicellulose degradation by rumen bacteria. Fed. Proc. 32:1819-1825.
17. Dehority, B. A. 1991. Effects of microbial synergism on fibre digestion in the rumen. Proc. Nutr. Soc. 50:149-159.
18. Dehority, B. A. 1993. Microbial ecology of cell wall fermentation. p 425. In H. G. Jung, D. R. Buxton, R. D. Hatfield and J. Ralph (ed.), Forage Cell Wall Structure and Digestibility, ASA-CSSA-SSSA, Madison, WI, USA.
19. Dehority, B. A. and C. G. Orpin. 1988. Development of, and natural fluctuations in, rumen microbial populations. p 151. In P. N. Hobson (ed.), The Rumen Microbial Ecosystem, Elsevier Applied Science, London.
20. Dehority, B. A. and H. W. Scott. 1967. Extent of cellulose and hemicellulose digestion in various forages by pure cultures of rumen bacteria. J. Dairy Sci. 50:1136-1141.
21. Dehority, B. A. and P. A. Tirabasso. 1993. Antibiosis between rumen bacteria and fungi. 22nd Biennial Conference on Rumen Function, Chicago, IL. November 9-11, p. 6.
22. Eadie, J. M. 1962. Interrelationships between certain rumen ciliate protozoa. J. Gen. Microbiol. 29:579-588.
23. Eadie, J. M. 1967. Studies on the ecology of certain rumen ciliate protozoa. J. Gen. Microbiol. 49:175-194.
24. Engles, F. M. and R. E. Brice. 1985. A barrier covering lignified cell walls of barley straw that restricts access by rumen microorganisms. Curr. Microbiol. 12:217-224.
25. Fondevila, M. and B. A. Dehority. 1994. Degradation and utilization of forage hemicellulose by rumen bacteria, singly in coculture or added sequentially. J. Appl. Bacteriol. 77:541-548.
26. Fondevila, M. and B. A. Dehority. 1996. Interactions between Fibrobacter succinogenes, Prevotella ruminicola and Ruminococcus flavefaciens in the digestion of cellulose from forages. J. Anim. Sci. 74:678-684.
27. Fonty, G., Ph. Gouet and V. Sante. 1988. Influence d'une bactérie méthanogène sur l'activité cellulolytique et al métabolisme de deux espèces de champignons cellulolytiques du rumen in vitro. Résultats préliminaires. Reprod. Nutr. Develop. 28:133-134.
28. Gradel, C. M. and B. A. Dehority. 1972. Fermentation of isolated pectin and pectin from intact forages by pure cultures of rumen bacteria. Appl. Microbiol. 23:332-340.
29. Gutierrez, J. 1955. Observations on bacterial feeding by the rumen ciliate Isotricha prostoma. J. Protozool. 5:122-126.
30. Gutierrez, J. and R. E. Davis. 1959. Bacterial ingestion by the rumen ciliates Entodinium and Diplodinium. J. Protozool. 6:222-226.
31. Gutierrez, J. and R. E. Hungate. 1957. Interrelationships between certain bacteria and the rumen ciliate Dasytricha ruminantium. Science 126:511.
32. Ho, Y. W., N. Abdullah and S. Jalaludin. 1988. Penetrating structures of anaerobic rumen fungi in cattle and swamp buffalo. J. Gen. Microbiol. 134:177-181.
33. Irvine, H. L. and C. S. Stewart. 1991. Interactions between anaerobic cellulolytic bacteria and fungi in the presence of Methanobrevibacter smithii. Lett. Appl. Microbiol. 12:62-64.
34. Joblin, K.N. and G. E. Naylor. 1993. Inhibition of the rumen anaerobic fungus Neocallimastix frontalis by fermentation products. Lett. Appl. Microbiol. 16:254-256.
35. Joblin, K. N. and A. G. Williams. 1991. Effect of cocultivation of ruminal chytrid fungi with Methanobrevi-bacter smithii on lucerne stem degradation and extracellular fungal enzyme activities. Lett. Appl. Microbiol. 12:121-124.
36. Joblin, K. N., G. Naylor and A. G. Williams. 1990. The effect of Methanobrevibacter smithii on the xylanolytic activity of anaerobic rumen fungi. Appl. Environ. Microbiol. 56:2287-2295.
37. Kalmokoff, M. L. and R. M. Teather. 1997. Isolation and characterization of a bacteriocin (Butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: evidence in support of the widespread occurrence of bacteriocin-like activity among rumen isolates of B. fibrisolvens. Appl. Environ. Microbiol. 63:394-402.
38. Kalmokoff, M. L., R. J. Forster and R. M. Teather. 1995. Production of bacteriocins among isolates of the rumen bacterium Butyrivibrio fibrisolvens. 1995 Dairy Research Report. University of Guelph Publication No. 0395. Ontario Ministry of Agriculture, Food and Rural Affairs. Guelph, Ontario, NIG 2W1, Canada. p 11-13.
39. Lowe, S. E., M. K. Theodorou and A.P.J. Trinci. 1987. Growth and fermentation of an anaerobic rumen fungus on various carbon sources and effect of temperature or development. Appl. Environ. Microbiol. 53:1210-1215.
40. Lubinsky, G. 1957. Note on the phylogenetic significance of predatory habits in the Ophryoscolecidae (Ciliata:Oligotricha). Can J. Zool. 35:579-580.
41. Miura, H., Horiguchi, M. & and T. Matsumoto. 1980. Nutritional interdependence among rumen bacteria, Bacteroides amylophilus, Megasphaera elsdenii, and Ruminococcus albus. Appl. Environ. Microbiol. 40:294-300.
42. Morris, E. J. and N. O. van Gylswyk. 1980 Comparison of the action of rumen bacteria on cells from Eragrostis tef. J. Agric. Sci. 95:313-323.
43. Mountfort, D. O., R. A. Asher and T. Bauchop. 1982. Fermentation of cellulose to methane and carbon dioxide by a rumen anaerobic fungus in a triculture with Methanobrevibacter sp. strain RA1 and Methanosarcina barkeri. Appl. Environ. Microbiol. 44:128-134.
44. Newbold, C. J. and K. Hillman. 1990. The effect of ciliate protozoa on the turnover of bacterial and fungal protein in the rumen of sheep. Lett. Appl. Microbiol. 11:100-102.
45. Odenyo, A. A., R. I. Mackie, D. A. Stahl and B. A. White. 1994. The use of 16S rDNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: develop ment of probes for Ruminococcus species and evidence for bacteriocin production. Appl. Environ. Microbiol. 60:3688-3696.
46. Odenyo, A. A., R. I. Mackie, D. A. Stahl and B. A. White. 1994. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: pure culture studies with cellulose and alkaline peroxide-treated wheat straw. Appl. Environ. Microbiol. 60:3697-3703.
47. Onodera R. and C. Henderson. 1980. Growth factors of bacterial origin for the culture of the rumen oligotrich protozoan, Entodinium caudatum. J. Appl. Bacteriol. 48:125-133.
48. Orpin, C. G. 1977. Studies on the defaunation of the ovine rumen using dioctyl sodium sulphosuccinate. J. Appl. Bacteriol. 43:309-318.
49. Osborne, J. M. and B. A. Dehority. 1989. Synergism in degradation and utilization of intact forage cellulose, hemicellulose, and pectin by three pure cultures of ruminal bacteria. Appl. Environ. Microbiol. 55:2247-2250.
50. Prins, R. A. 1991. The rumen ciliates and their functions, p 39-52. In J. P. Jouany (ed.), Rumen Microbial Metabolism and Ruminant Digestion, INRA Editions, Paris.
51. Prins, R. A. and C.J.A.H.V. van den Vorstenbosch. 1975. Interrelationships between rumen microorganisms, p 15. In Miscellaneous Papers, Landbouwhogeschool Wageningen.
52. Roger, V., A. Bernalier, E. Grenet, G. Fonty, J. Jamot and P. Gouet. 1993. Degradation of wheat straw and maize stem by a monocentric and a polycentric rumen fungi alone or in association with rumen cellulolytic bacteria. Anim. Feed Sci. Technol. 42:69-82.
53. Romulo, B. H., S. H. Bird and R. A. Leng. 1986. The effects of defaunation on digestibility and rumen fungi counts in sheep fed high-fibre diets. Proc. Aust. Soc. Anim. Prod. 16:327-330.
54. Russell, J. B. 1985. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl. Environ. Microbiol. 49:572-576.
55. Russell, J. B. and R. J. Wallace. 1988. Energy yielding and consuming reactions. pp 185-216. In P. N. Hobson (ed.), The Rumen Microbial Ecosystem. London: Elsevier Science Publications Ltd.
56. Saluzzi, L., A. Smith and C. S. Stewart. 1993. Analysis of bacterial phospholipid markers and plant monosaccharides during forage degradation by Ruminococcus flavefaciens and Fibrobacter succinogenes in co-culture. J. Gen. Microbiol. 139:2865-2873.
57. Scheifinger, C. C. and M. J. Wolin. 1973. Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 25:789-795.
58. Soetanto, H., G.L.R. Gordon, I. D. Hume and R. A. Leng. 1985. The role of protozoa and fungi in fibre digestion in the rumen of sheep. Proceedings of the 3rd AAAP Animal Science Congress 2:805-807.
59. Stewart, C. S., S. H. Duncan, A. J. Richardson, C. Backwell and R. Begbie. 1992. The inhibition of fungal cellulolysis by cell-free preparations from ruminococci. FEMS Microbiol. Lett. 97:83-88.
60. Ushida, K., H. Tanaka and Y. Kojima. 1989. A simple in situ method for estimating fungal population size in the rumen. Lett. Appl. Microbiol. 9:109-111.
61. Williams, A. G. and G. S. Coleman. 1988. The rumen protozoa, p 77-128. In P. N. Hobson (ed.), The Rumen Microbial Ecosystem, Elsevier Applied Science, London.
62. Williams, A. G. and G. S. Coleman. 1992. The rumen protozoa. Springer-Verlag New York, Inc. New York, N.Y.
63. Williams, A. G. and S. E. Withers. 1993. Changes in the rumen microbial populations and its activities during the refaunation period after the reintroduction of ciliate protozoa into the rumen of defaunated sheep. Can. J. Microbiol. 31:61-69.
64. Williams, A. G., K. N. Joblin and G. Fonty. 1994. Interactions between the rumen chytrid fungi and other microorganisms, p. 191-228. In D. O. Mountfort and C. G. Orpin (ed.), Anaerobic Fungi, Marcel Dekker, Inc., New York.
65. Williams, A. G., S. E. Withers and K. N. Joblin. 1994. The effect of cocultivation with hydrogen-consuming bacteria or xylanolysis by Ruminococcus flavefaciens. Curr. Microbiol. 29:133-138.
66. Wolin, M. J. and T. L. Miller. 1988. Microbe-microbe interactions. pp 343-359. In P. N. Hobson (ed.), The Rumen Microbial Ecosystem. Elsevier Sci. Publ. Ltd. London.